CeSTAAN: C. elegans Single nucleus Transcriptomic Atlas of Adult Neurons
Welcome to Our Adult Single Nucleus RNA Sequencing Data
Overview of This Site
The data on this website and behavioral analysis and findings come from our publications:
St. Ange and Weng et al. Cell Genomics 2024.Morillo et al. Cell Reports 2025.- What is our dataset? We developed a protocol to isolate and sequence individual neuronal nuclei from adult C. elegans. Here we provide an easy way to access our high resolution transcriptomic data for adult animals' neurons.
- Why single nucleus instead of single cell? In certain tissue types, the cell isolation protocol causes shearing and breakage of the desired cells. In neurons, this includes axon breakage and cell leakage. This makes it difficult to capture synaptically localized mRNA transcripts for example.
- What genotypes have you sequenced? We have sequenced Adult Day 1 N2 and daf-2 animals as well as adult males and genotypically and age matched hermaphrodites.
Query
Query
Generate UMAP by Gene
Enter one or two genes to visuzalize their expression on a UMAP plot.
Note: plot genertion takes 1 to 2 minutes
Heatmap generation
Note: The generated heatmap is dynamic, you may need to zoom in to see all neurons on the x-axis
Violin plot generation
Plot generation takes 1 to 2 minutes
Note: When showing differential expression it is N2 vs daf-2 or Male vs Hermaphrodite
Seurat Object
The single nucleus data is an .rds file containing Seurat objects for all cells. This website contains many optimized queries for interfacing with the data, but if your analysis is more fine-grained, you may download the 7GB object below.
Download WT/daf-2 DataDownload Male data
snSeq Papers by the Murphy Lab
There are various circumstances where single nucleus RNA sequencing is the appropriate way to ask a biological question. Here are some papers by our lab where we've employed this method.

Gene expression in individual neurons can change during development to adulthood and can have large effects on behavior. Additionally, the insulin/insulin-like signaling (IIS) pathway regulates many of the adult functions of Caenorhabditis elegans, including learning and memory, via transcriptional changes. We used the deep resolution of single-nucleus RNA sequencing to define the adult transcriptome of each neuron in wild-type and daf-2 mutants, revealing expression differences between L4 larval and adult neurons in chemoreceptors, synaptic genes, and learning/memory genes. We used these data to identify adult new AWC-specific regulators of chemosensory function that emerge upon adulthood. daf-2 gene expression changes correlate with improved cognitive functions, particularly in the AWC sensory neuron that controls learning and associative memory; behavioral assays of AWC-specific daf-2 genes revealed their roles in cognitive function. Combining technology and functional validation, we identified conserved genes that function in specific adult neurons to control behavior, including learning and memory.
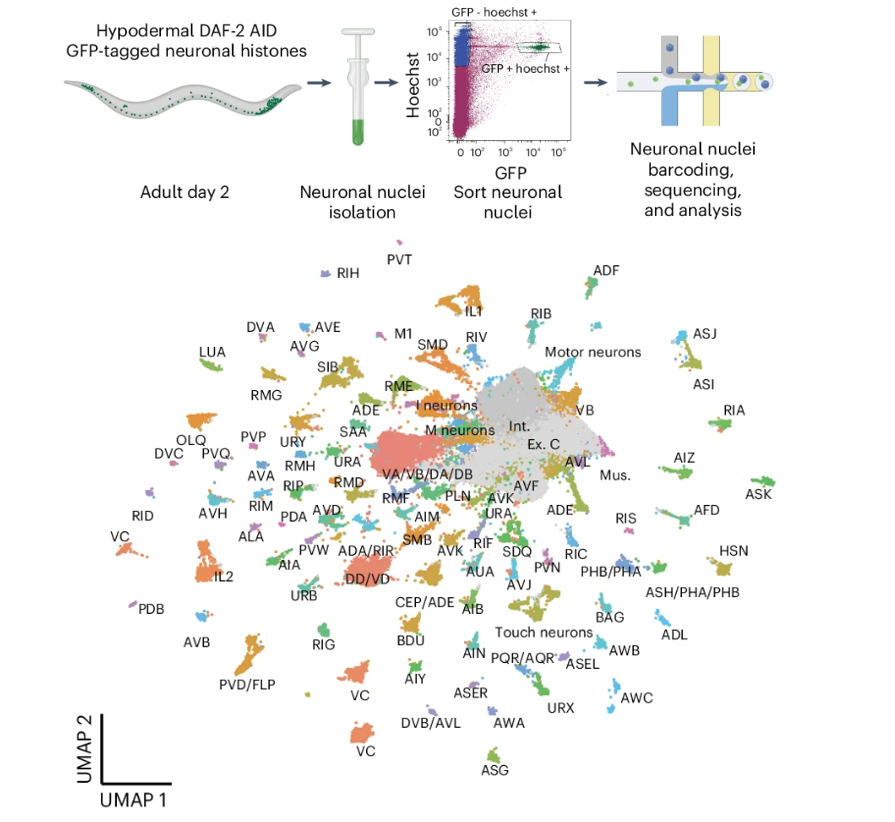
While memory regulation is predominantly understood as autonomous to neurons, factors outside the brain can also affect neuronal function. In Caenorhabditis elegans, the insulin/IGF-1-like signaling (IIS) pathway regulates longevity, metabolism and memory: long-lived daf-2 insulin/IGF-1 receptor mutants more than double memory duration after a single training session, and it was assumed that memory regulation was strictly neuronal. However, here we show that degradation of DAF-2 in the hypodermis also greatly extends memory, via expression of the diffusible Notch ligand, OSM-11, which in turn activates Notch signaling in neurons. Single-nucleus RNA sequencing of neurons revealed increased expression of CREB and other memory genes. Furthermore, in aged animals, activation of the hypodermal IIS–Notch pathway as well as OSM-11 overexpression rescue both memory and learning via CREB activity. Thus, insulin signaling in the liver-like hypodermis non-autonomously regulates neuronal function, providing a systemic connection between metabolism and memory through IIS–Notch–CREB signaling from the body to the brain.
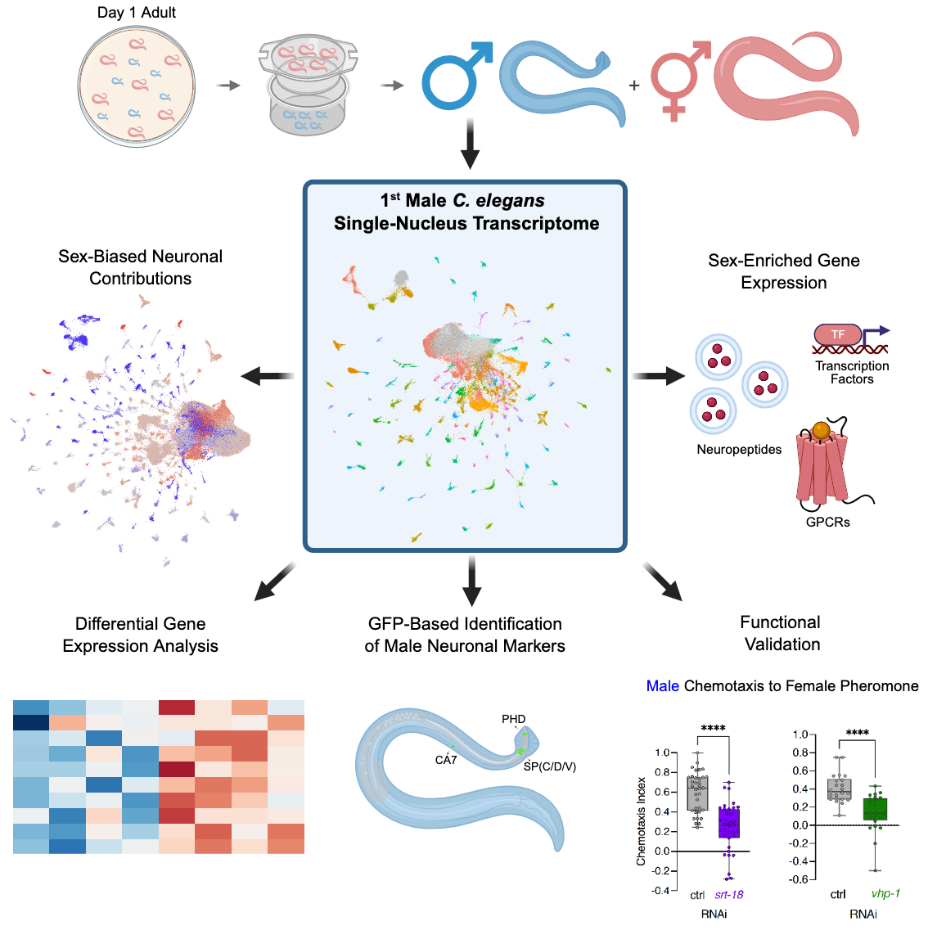
Sexual differentiation of the nervous system causes differences in neuroanatomy, synaptic connectivity, and physiology. These sexually-dimorphic phenotypes ultimately translate into profound behavioral differences. C. elegans’ two sexes, XO males and XX hermaphrodites, demonstrate differences in neurobiology and behavior. However, the neuron class and sex-specific transcriptomic differences, particularly at the single-neuron level, that cause such phenotypic divergence remains understudied. Here, using single-nucleus RNA sequencing, we assessed and compared adult male and hermaphrodite C. elegans neuronal transcriptomes, identifying sex-specific neurons, including previously-unannotated male neurons. Sex-shared neurons displayed large expression differences, with some neuron classes clustering as distinct neurons between the sexes. Males express ∼100 male-specific GPCRs, largely limited to a subset of neurons. We identified the most highly-divergent neurons between the sexes, and functionally characterized a sex-shared target, vhp-1, in male-specific pheromone chemotaxis. Our data provide a resource for discovering nervous-system-wide sex transcriptomic differences and the molecular basis of sex-specific behaviors.